Search Result
Results for "
base pair
" in MedChemExpress (MCE) Product Catalog:
2
Biochemical Assay Reagents
| Cat. No. |
Product Name |
Target |
Research Areas |
Chemical Structure |
-
- HY-148424
-
|
|
ADC Payload
DNA Alkylator/Crosslinker
|
Cancer
|
|
PBD dimer-2 (compound 2c) is a C8-linked pyrrolobenzodiazepine dimer. PBD dimer-2 can span an extra base pair and cross-link the 5′-Pu-GA(T/A)TC-Py sequence. PBD dimer-2 can be used as a payload for antibody–agent conjugates (ADCs), and it can be used for the research of cancer .
|
-

-
- HY-119840
-
|
|
DNA/RNA Synthesis
|
Cancer
|
|
Anthragallol can insert base pairs of DNA. Anthragallol exhibits cytotoxicity by binding to DNA .
|
-

-
- HY-W006395
-
-

-
- HY-D0814
-
|
4',6-Diamidino-2-phenylindole dihydrochloride
|
DNA Stain
|
Others
|
|
DAPI dihydrochloride is a DAPI dye. DAPI is a fluorescent dye that binds strongly to DNA. It binds to the AT base pair of the double-stranded DNA minor groove, and one DAPI molecule can occupy three base pair positions. The fluorescence intensity of DAPI molecules bound to double-stranded DNA is increased by about 20 times, and it is commonly observed with fluorescence microscopy, and the amount of DNA can be determined based on the intensity of fluorescence. In addition, because DAPI can pass through intact cell membranes, it can be used to stain both live and fixed cells .
|
-

-
- HY-D1396
-
Br-DAPI
3 Publications Verification
|
Fluorescent Dye
DNA Stain
|
Others
|
|
Br-DAPI is a marker dye in DAPI series. DAPI is a fluorescent dye that binds strongly to DNA. It binds to the AT base pair of the double-stranded DNA minor groove, and one DAPI molecule can occupy three base pair positions. The fluorescence intensity of DAPI molecules bound to double-stranded DNA is increased by about 20 times, and it is commonly observed with fluorescence microscopy, and the amount of DNA can be determined based on the intensity of fluorescence. In addition, because DAPI can pass through intact cell membranes, it can be used to stain both live and fixed cells . Storage: Keep away from light.
|
-

-
- HY-W002272
-
|
|
Nucleoside Antimetabolite/Analog
|
Others
|
|
Isocytosine is a non-natural nucleobase and an isomer of cytosine. It is used in combination with Isoguanine in studies of unnatural nucleic acid analogues of the normal base pairs in DNA and used as a nucleobase of hachimoji RNA .
|
-

-
- HY-21997
-
|
|
DNA/RNA Synthesis
Drug Intermediate
|
Others
|
|
DMT-2'fluoro-da(bz) amidite is a key intermediate for synthesizing antisense oligonucleotides with high nuclease resistance, high RNA binding affinity, and maintained base-pair specificity .
|
-

-
- HY-D0814A
-
|
4',6-Diamidino-2-phenylindole dihydrochloride (solution)
|
DNA Stain
|
Others
|
|
DAPI dihydrochloride (4',6-Diamidino-2-phenylindole dihydrochloride) (solution) is a DAPI dye. DAPI is a fluorescent dye that can bind strongly to DNA. DAPI binds to the A/T base pairs in the minor groove of double-stranded DNA (1 DAPI molecule can occupy 3 base pairs), which amplifies the fluorescence intensity by about 20 times. By observing the fluorescence intensity, DNA can be quantified. DAPI can also penetrate intact cell membranes and is often used to stain living and fixed cells .
|
-

-
- HY-157549A
-
|
AYX1 sodium
|
Nucleoside Antimetabolite/Analog
|
Neurological Disease
|
|
Brivoligide (AYX1) sodium is a double-stranded, unprotected, 23 base-pair oligonucleotide. Brivoligide sodium can reduce acute post-surgical pain. Brivoligide sodium mimics the DNA sequence normally bound by EGR1 on chromosomes .
|
-

-
- HY-W006395R
-
-

-
- HY-50071
-
-

-
- HY-157549
-
|
AYX1
|
Nucleoside Antimetabolite/Analog
|
Neurological Disease
|
|
Brivoligide (AYX1) is a double-stranded, unprotected, 23 base-pair oligonucleotide. Brivoligide can reduce acute post-surgical pain. Brivoligide mimics the DNA sequence normally bound by EGR1 on chromosomes .
|
-

-
- HY-D1079
-
|
|
DNA Stain
|
Others
|
|
EDANS sodium is a potent fluorogenic substrates. EDANS sodium is a donor for FRET-based nucleic acid probes and protease substrates. EDANS sodium is often paired with DABCYL or DABSYL. The optimal absorbance and emission wavelengths of EDANS sodium are λabs = 336 nm and λem = 490 nm respectively .
|
-

-
- HY-117813
-
|
|
Nucleoside Antimetabolite/Analog
DNA/RNA Synthesis
Dengue Virus
Flavivirus
CHIKV
SARS-CoV
|
Infection
|
|
2-Thiouridine is an orally active modified nucleobase. 2-Thiouridine stabilizes U:A base pairs and destabilizes U:G wobble base pairs. 2-Thiouridine significantly improves the efficiency and accuracy of nonenzymatic replication of mixed-sequence A/U-containing RNA templates. 2-Thiouridine exhibits antiviral activity against multiple positive-sense single-stranded RNA viruses (DENV2, ZIKV, YFV, JEV, WNV, CHIKV, human coronaviruses ( [HCoV]-229E, HCoV-OC43, SARS-CoV), and MERS-CoV) .
|
-

-
- HY-114577
-
|
Isophosphoramide mustard tromethamine; IPM tromethamine; ZIO-201 tromethamine
|
DNA Alkylator/Crosslinker
|
Cancer
|
|
Palifosfamide (tromethamine) is a synthetic alkylating agent with potential antineoplastic activity. As the stabilized active metabolite of ifosfamide, palifosfamide (tromethamine) irreversibly alkylates and crosslinks DNA through GC base pairs. This leads to an inhibition of DNA replication and ultimately cell death. Compared to ifosfamide, palifosfamide (tromethamine) is less toxic.
|
-

-
- HY-162819
-
|
|
Apoptosis
|
Cancer
|
|
Apoptosis inducer 26 (compound [AgCl(dap2SH)(PPh3)2]) is an autophagy inducer based on mononuclear Ag(I) ligands, with antibacterial and anticancer activities against a variety of bacterial strains and cancer cell lines. Apoptosis inducer 26 can effectively inhibit the growth of both Gram(+) and Gram(-) bacteria by causing the accumulation of Ag(I) ions in the bacterial periplasm. Apoptosis inducer 26 can intercalate between base pairs of CT DNA and induce apoptosis in A549 cells. Apoptosis inducer 26 also has the ability to scavenge free radicals, which can protect against oxidative stress .
|
-

-
- HY-12404
-
|
Diminazene diaceturate
|
Parasite
Angiotensin-converting Enzyme (ACE)
|
Infection
Inflammation/Immunology
|
|
Diminazene aceturate (Diminazene diaceturate) is an anti-trypanosome agent for livestock. The main biochemical mechanism of the trypanocidal actions of Diminazene aceturate is by binding to trypanosomal kinetoplast DNA (kDNA) in a non-intercalative manner through specific interaction with sites rich in adenine-thymine base pairs. Diminazene aceturate is also an angiotensin-converting enzyme 2 (ACE2) activator and has strong and potent anti-inflammatory properties .
|
-

-
- HY-113138
-
|
N3-Methyluridine
|
Endogenous Metabolite
|
Metabolic Disease
|
|
3-Methyluridine (m 3U; N3-Methyluridine) is a methylated nucleotide present in ribosomal RNA (rRNA), mainly targeting specific base sites of RNA molecules such as 23S rRNA. 3-Methyluridine can introduce a methyl group at the N3 position of uracil, affecting the secondary structure stability and base pairing ability of RNA, and regulating ribosome function. For example, it affects ribosomal subunit binding and tRNA interaction. 3-Methyluridine is often used as a key raw material for the synthesis of modified nucleotides, and is used to construct RNA oligonucleotides containing methylation modifications to study the effects of RNA methylation on gene expression and drug resistance .
|
-

-
- HY-W009444
-
|
|
Endogenous Metabolite
|
Others
|
|
5-Methyluridine (m 5U) is an RNA modified nucleotide generated by RNA methyltransferases (such as TrmA and RumA), which mainly targets specific uracil sites in RNA molecules such as the T arm of tRNA and rRNA. 5-Methyluridine relies on enzyme recognition of RNA secondary/tertiary structures (such as the T loop of tRNA or the specific stem-loop structure of rRNA) and participates in physiological processes such as translation accuracy and ribosome function by stabilizing RNA folding or regulating base pairing .
|
-

-
- HY-12404R
-
|
Diminazene diaceturate (Standard)
|
Reference Standards
Parasite
Angiotensin-converting Enzyme (ACE)
|
Infection
Inflammation/Immunology
|
|
Diminazene (aceturate) (Standard) is the analytical standard of Diminazene (aceturate). This product is intended for research and analytical applications. Diminazene aceturate (Diminazene diaceturate) is an anti-trypanosome agent for livestock. The main biochemical mechanism of the trypanocidal actions of Diminazene aceturate is by binding to trypanosomal kinetoplast DNA (kDNA) in a non-intercalative manner through specific interaction with sites rich in adenine-thymine base pairs. Diminazene aceturate is also an angiotensin-converting enzyme 2 (ACE2) activator and has strong and potent anti-inflammatory properties .
|
-

-
- HY-113138R
-
|
|
Endogenous Metabolite
|
Metabolic Disease
|
|
3-Methyluridine (Standard) is the analytical standard of 3-Methyluridine. This product is intended for research and analytical applications. 3-Methyluridine (m3U; N3-Methyluridine) is a methylated nucleotide present in ribosomal RNA (rRNA), mainly targeting specific base sites of RNA molecules such as 23S rRNA. 3-Methyluridine can introduce a methyl group at the N3 position of uracil, affecting the secondary structure stability and base pairing ability of RNA, and regulating ribosome function. For example, it affects ribosomal subunit binding and tRNA interaction. 3-Methyluridine is often used as a key raw material for the synthesis of modified nucleotides, and is used to construct RNA oligonucleotides containing methylation modifications to study the effects of RNA methylation on gene expression and drug resistance .
|
-

-
- HY-173192
-
|
|
Bacterial
|
Infection
|
|
Antibacterial agent 272 (Compound Z22) is a potential antimicrobial agent targeting DNA and the DNA-topoisomerase II (DNA-Topo II) complex, exhibiting MIC values of 1 μg/mL against Staphylococcus aureus 25923 and 29213, 2 μg/mL against Staphylococcus epidermidis 12228, 2-4 μg/mL against Enterococcus faecalis, and 4 μg/mL against Pseudomonas aeruginosa 9027 and 27853, demonstrating potent antibacterial activity. This compound functions by intercalating with DNA base pairs to disrupt normal bacterial DNA function, making it suitable for research on bacterial infectious diseases .
|
-
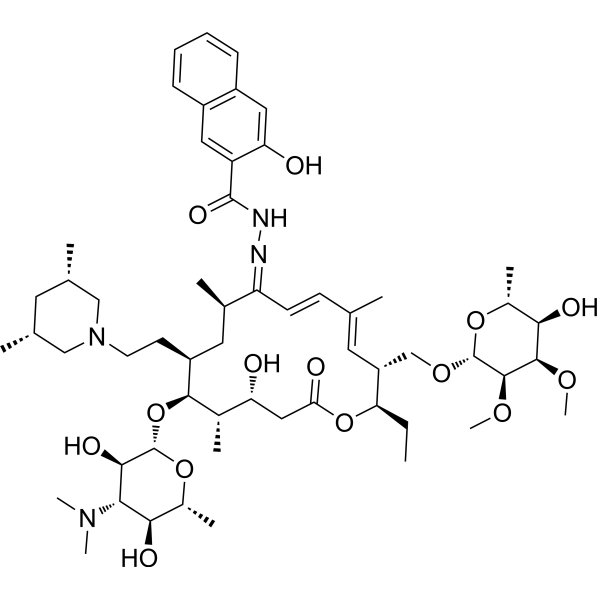
-
- HY-170557
-
|
|
Topoisomerase
Apoptosis
Mitochondrial Metabolism
|
Cancer
|
|
Topoisomerase IIα-IN-10 (Compound 13r) is an inhibitor of Topoisomerase IIα. It binds to the active site of DNA when complexed with Topoisomerase IIα, and this binding is stabilized through interactions with DNA base pairs and amino acid residues. Topoisomerase IIα-IN-10 can induce Apoptosis by intercalating DNA and inhibiting Topoisomerase IIα, thereby disrupting the mitochondrial membrane potential and inhibiting the growth of HCT116 cell lines, with an IC50 of 4.37 μM against HCT116 cells. Topoisomerase IIα-IN-10 can be used for research in the field of cancer treatment .
|
-

-
- HY-D0917
-
|
|
DNA Stain
|
Cancer
|
|
TO-PRO 1 is a DNA-binding fluorescent dye for non-living cells (Ex/Em=515 nm/531 nm). TO-PRO 1 can intercalate into base pairs of double-stranded DNA and produce stronger fluorescence. TO-PRO 1 is suitable for necrotic cells or late apoptotic cells with damaged cell membranes, showing green fluorescence under fluorescence microscopy or flow cytometry. TO-PRO 1 can be used to distinguish live cells from dead cells and distinguish cell membrane integrity. TO-PRO 1 can be attached to the surface of Feraheme (FH) nanoparticles (NPs) to obtain fluorescent dye-functionalized NPs for drug delivery studies .
|
-

-
- HY-153843
-
|
|
Biochemical Assay Reagents
|
Others
|
|
RNA Aptamer Corn (sodium) is a 28-nt-long aptamer that is substantially shorter than Spinach and Spinach2 and exhibits bright red fluorescence upon binding DFHO (a soluble analog of the intrinsic fluorophore of red fluorescent protein), RNA Aptamer Corn (sodium) can be used to visualize RNA expression or localization in live cells which have been soaked with chromophores. The Corn-DFHO does not become appreciably cytotoxic when illuminated. And most importantly, Corn-DFHO exhibits markedly increased photostability compared to other aptamer-chromophore complexes both in vitro and in vivo. (36 nt Corn construct: 5'-GGCGCGAGGAAGGAGGUCUGAGGAGGUCACUGCGCC-3'; A 36-nt RNA construct, comprised of the 28-nt minimal Corn sequence extended proximally with a 4 base-pair stem.)
|
-

| Cat. No. |
Product Name |
Type |
-
- HY-D0814
-
|
4',6-Diamidino-2-phenylindole dihydrochloride
|
Fluorescent Dyes/Probes
|
|
DAPI dihydrochloride is a DAPI dye. DAPI is a fluorescent dye that binds strongly to DNA. It binds to the AT base pair of the double-stranded DNA minor groove, and one DAPI molecule can occupy three base pair positions. The fluorescence intensity of DAPI molecules bound to double-stranded DNA is increased by about 20 times, and it is commonly observed with fluorescence microscopy, and the amount of DNA can be determined based on the intensity of fluorescence. In addition, because DAPI can pass through intact cell membranes, it can be used to stain both live and fixed cells .
|
-
- HY-D0814A
-
|
4',6-Diamidino-2-phenylindole dihydrochloride (solution)
|
Fluorescent Dyes/Probes
|
|
DAPI dihydrochloride (4',6-Diamidino-2-phenylindole dihydrochloride) (solution) is a DAPI dye. DAPI is a fluorescent dye that can bind strongly to DNA. DAPI binds to the A/T base pairs in the minor groove of double-stranded DNA (1 DAPI molecule can occupy 3 base pairs), which amplifies the fluorescence intensity by about 20 times. By observing the fluorescence intensity, DNA can be quantified. DAPI can also penetrate intact cell membranes and is often used to stain living and fixed cells .
|
-
- HY-D0917
-
|
|
DNA Stain
|
|
TO-PRO 1 is a DNA-binding fluorescent dye for non-living cells (Ex/Em=515 nm/531 nm). TO-PRO 1 can intercalate into base pairs of double-stranded DNA and produce stronger fluorescence. TO-PRO 1 is suitable for necrotic cells or late apoptotic cells with damaged cell membranes, showing green fluorescence under fluorescence microscopy or flow cytometry. TO-PRO 1 can be used to distinguish live cells from dead cells and distinguish cell membrane integrity. TO-PRO 1 can be attached to the surface of Feraheme (FH) nanoparticles (NPs) to obtain fluorescent dye-functionalized NPs for drug delivery studies .
|
| Cat. No. |
Product Name |
Type |
-
- HY-21997
-
|
|
Gene Sequencing and Synthesis
|
|
DMT-2'fluoro-da(bz) amidite is a key intermediate for synthesizing antisense oligonucleotides with high nuclease resistance, high RNA binding affinity, and maintained base-pair specificity .
|
-
- HY-117813
-
|
|
Gene Sequencing and Synthesis
|
|
2-Thiouridine is an orally active modified nucleobase. 2-Thiouridine stabilizes U:A base pairs and destabilizes U:G wobble base pairs. 2-Thiouridine significantly improves the efficiency and accuracy of nonenzymatic replication of mixed-sequence A/U-containing RNA templates. 2-Thiouridine exhibits antiviral activity against multiple positive-sense single-stranded RNA viruses (DENV2, ZIKV, YFV, JEV, WNV, CHIKV, human coronaviruses ( [HCoV]-229E, HCoV-OC43, SARS-CoV), and MERS-CoV) .
|
| Cat. No. |
Product Name |
Category |
Target |
Chemical Structure |
-
- HY-119840
-
-

-
- HY-W002272
-
-

-
- HY-117813
-
|
|
Source classification
|
Nucleoside Antimetabolite/Analog
DNA/RNA Synthesis
Dengue Virus
Flavivirus
CHIKV
SARS-CoV
|
|
2-Thiouridine is an orally active modified nucleobase. 2-Thiouridine stabilizes U:A base pairs and destabilizes U:G wobble base pairs. 2-Thiouridine significantly improves the efficiency and accuracy of nonenzymatic replication of mixed-sequence A/U-containing RNA templates. 2-Thiouridine exhibits antiviral activity against multiple positive-sense single-stranded RNA viruses (DENV2, ZIKV, YFV, JEV, WNV, CHIKV, human coronaviruses ( [HCoV]-229E, HCoV-OC43, SARS-CoV), and MERS-CoV) .
|
-

-
- HY-113138
-
|
N3-Methyluridine
|
Natural Products
Classification of Application Fields
Source classification
Other Diseases
Endogenous metabolite
Disease Research Fields
|
Endogenous Metabolite
|
|
3-Methyluridine (m 3U; N3-Methyluridine) is a methylated nucleotide present in ribosomal RNA (rRNA), mainly targeting specific base sites of RNA molecules such as 23S rRNA. 3-Methyluridine can introduce a methyl group at the N3 position of uracil, affecting the secondary structure stability and base pairing ability of RNA, and regulating ribosome function. For example, it affects ribosomal subunit binding and tRNA interaction. 3-Methyluridine is often used as a key raw material for the synthesis of modified nucleotides, and is used to construct RNA oligonucleotides containing methylation modifications to study the effects of RNA methylation on gene expression and drug resistance .
|
-

-
- HY-W009444
-
-

-
- HY-113138R
-
|
|
Natural Products
Source classification
Endogenous metabolite
|
Endogenous Metabolite
|
|
3-Methyluridine (Standard) is the analytical standard of 3-Methyluridine. This product is intended for research and analytical applications. 3-Methyluridine (m3U; N3-Methyluridine) is a methylated nucleotide present in ribosomal RNA (rRNA), mainly targeting specific base sites of RNA molecules such as 23S rRNA. 3-Methyluridine can introduce a methyl group at the N3 position of uracil, affecting the secondary structure stability and base pairing ability of RNA, and regulating ribosome function. For example, it affects ribosomal subunit binding and tRNA interaction. 3-Methyluridine is often used as a key raw material for the synthesis of modified nucleotides, and is used to construct RNA oligonucleotides containing methylation modifications to study the effects of RNA methylation on gene expression and drug resistance .
|
-

| Cat. No. |
Product Name |
|
Classification |
-
- HY-21997
-
|
|
|
Nucleoside Phosphoramidites
Adenine
|
|
DMT-2'fluoro-da(bz) amidite is a key intermediate for synthesizing antisense oligonucleotides with high nuclease resistance, high RNA binding affinity, and maintained base-pair specificity .
|
-
- HY-W009444
-
|
|
|
Nucleoside Analogs
Uridine
|
|
5-Methyluridine (m 5U) is an RNA modified nucleotide generated by RNA methyltransferases (such as TrmA and RumA), which mainly targets specific uracil sites in RNA molecules such as the T arm of tRNA and rRNA. 5-Methyluridine relies on enzyme recognition of RNA secondary/tertiary structures (such as the T loop of tRNA or the specific stem-loop structure of rRNA) and participates in physiological processes such as translation accuracy and ribosome function by stabilizing RNA folding or regulating base pairing .
|
-
- HY-153843
-
|
|
|
Aptamers
|
|
RNA Aptamer Corn (sodium) is a 28-nt-long aptamer that is substantially shorter than Spinach and Spinach2 and exhibits bright red fluorescence upon binding DFHO (a soluble analog of the intrinsic fluorophore of red fluorescent protein), RNA Aptamer Corn (sodium) can be used to visualize RNA expression or localization in live cells which have been soaked with chromophores. The Corn-DFHO does not become appreciably cytotoxic when illuminated. And most importantly, Corn-DFHO exhibits markedly increased photostability compared to other aptamer-chromophore complexes both in vitro and in vivo. (36 nt Corn construct: 5'-GGCGCGAGGAAGGAGGUCUGAGGAGGUCACUGCGCC-3'; A 36-nt RNA construct, comprised of the 28-nt minimal Corn sequence extended proximally with a 4 base-pair stem.)
|
Your information is safe with us. * Required Fields.
Inquiry Information
- Product Name:
- Cat. No.:
- Quantity:
- MCE Japan Authorized Agent: